tna:
An R package for Transition Network Analysis
tna:
An R package for Transition Network Analysis

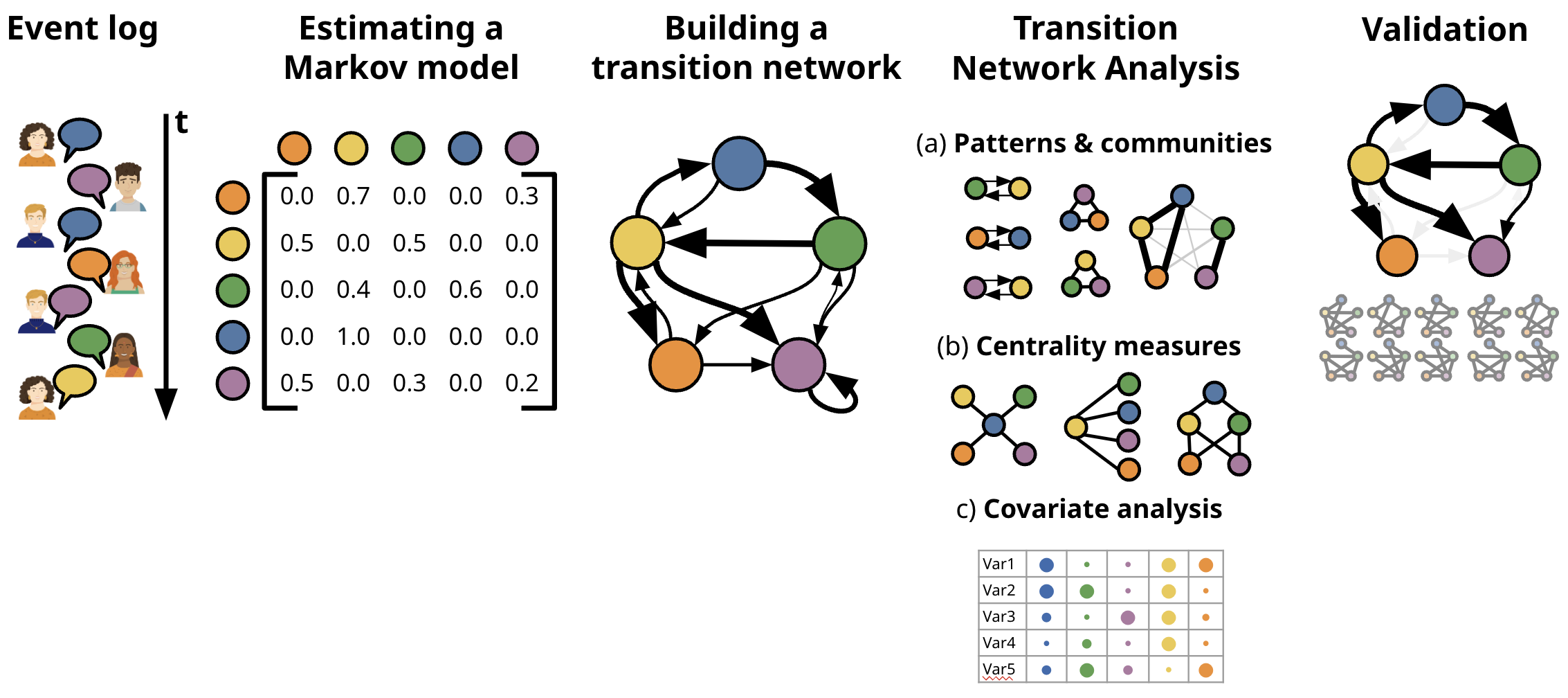
tna is an R package for the analysis of relational
dynamics through Transition Network Analysis (TNA). TNA provides tools
for building TNA models, plotting transition networks, calculating
centrality measures, and identifying dominant events and patterns. TNA
statistical techniques (e.g., bootstrapping and permutation tests)
ensure the reliability of observed insights and confirm that identified
dynamics are meaningful. See (Saqr et al., 2025)
for more details on TNA.
Check out our tutorials:
You can also try our Shiny app.
You can install the most recent stable version of tna
from CRAN or the
development version from GitHub by
running one of the following:
install.packages("tna")
# install.packages("devtools")
# devtools::install_github("sonsoleslp/tna")Load the package
library("tna")Example data
data("group_regulation", package = "tna")Build a Markov model
tna_model <- tna(group_regulation)summary(tna_model)| metric | value |
|---|---|
Plot the transition network
# Default plot
plot(tna_model) # Optimized plot
plot(
tna_model, cut = 0.2, minimum = 0.05,
edge.label.position = 0.8, edge.label.cex = 0.7
)
Calculate the centrality measures
cent <- centralities(tna_model)| state | OutStrength | InStrength | ClosenessIn | ClosenessOut | Closeness | Betweenness | BetweennessRSP | Diffusion | Clustering |
|---|---|---|---|---|---|---|---|---|---|
Plot the centrality measures
plot(cent, ncol = 3)Estimate centrality stability
estimate_centrality_stability(tna_model)
#> Centrality Stability Coefficients
#>
#> InStrength OutStrength Betweenness
#> 0.9 0.9 0.7Identify and plot communities
coms <- communities(tna_model)
plot(coms)Find and plot cliques
cqs <- cliques(tna_model, threshold = 0.12)
plot(cqs)Compare high achievers (first 1000) with low achievers (last 1000)
tna_model_start_high <- tna(group_regulation[1:1000, ])
tna_model_start_low <- tna(group_regulation[1001:2000, ])
comparison <- permutation_test(
tna_model_start_high,
tna_model_start_low,
measures = c("InStrength")
)Simple comparison vs. permutation test comparison
plot_compare(tna_model_start_high, tna_model_start_low)
plot(comparison)Compare centralities
print(comparison$centralities$stats)| state | centrality | diff_true | effect_size | p_value |
|---|---|---|---|---|