The goal of psc is to compare a dataset of observations against a parametric model
You can install the development version of psc from GitHub with:
# install.packages("devtools")
devtools::install_github("richJJackson/psc")This is a basic example which shows you how to solve a common problem:
library(psc)
#> Loading required package: survival
#> Loading required package: ggplot2
library(survival)
## basic example code
### Load model
data("surv.mod")
### Load Data
data("data")
#> Warning in data("data"): data set 'data' not found
### Use 'pscfit' to compare
surv.psc <- pscfit(surv.mod,data)
#> Warning in pscData_match(CFM$cov_class, CFM$cov_lev, DCcov): vi specified as a character in the model, consider respecifying
#> as a factor to ensure categories match between CFM and DC
#> Warning in pscData_match(CFM$cov_class, CFM$cov_lev, DCcov): allmets specified as a character in the model, consider respecifying
#> as a factor to ensure categories match between CFM and DCYou can use standard commands for getting a summary of your analysis…
summary(surv.psc)
#> Counterfactual Model (CFM):
#> A model of class 'flexsurvreg'
#> Fit with 3 internal knots
#>
#> CFM Formula:
#> Surv(time, cen) ~ vi/age60 + ecog + allmets + logafp + alb +
#> logcreat + logast + aet
#> <environment: 0x116b6fd60>
#>
#> CFM Summary:
#> Expected response for the outcome under the CFM:
#> S lo hi
#> 9.694 9.094 11.013
#>
#> Observed outcome from the Data Cohort:
#> [,1]
#> median 6.366
#> 0.95LCL 5.436
#> 0.95UCL 9.094
#>
#> MCMC Fit:
#> Posterior Distribution obtaine with fit summary:
#> variable rhat ess_bulk ess_tail mcse_mean
#> [1,] beta_1 1.001575 1157.117 1022.026 0.002900963
#>
#> Summary:
#> Posterior Distribution for beta:Call:
#> CFM model + beta
#>
#> Coefficients:
#> variable mean sd median q5
#> posterior beta_1 0.3707433 0.09903147 0.3708735 0.2103505
#> q95
#> posterior 0.5420673… and to see a plot of what you have done
#> variable mean sd median q5
#> posterior beta_1 0.3707433 0.09903147 0.3708735 0.2103505
#> q95
#> posterior 0.5420673
#> Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
#> ℹ Please use `linewidth` instead.
#> ℹ The deprecated feature was likely used in the ggpubr package.
#> Please report the issue at <https://github.com/kassambara/ggpubr/issues>.
#> This warning is displayed once every 8 hours.
#> Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
#> generated.
#> Ignoring unknown labels:
#> • colour : "Strata"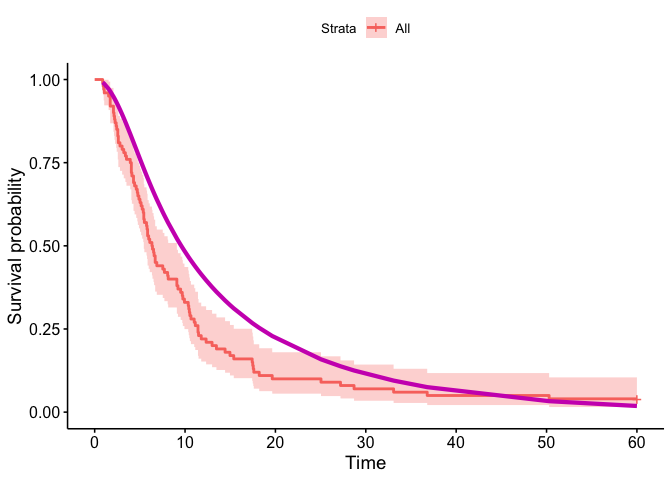
In that case, don’t forget to commit and push the resulting figure files, so they display on GitHub and CRAN.