Marlon E. Cobos, Hannah L. Owens, Jorge Soberón, A. Townsend Peterson
The package mop contains a set of tools to perform the
Mobility Oriented-Parity (MOP) metric, which helps to compare a set of
conditions of reference versus another set of of interest. The main
goals of the MOP metric are to explore conditions in the set of interest
that are non-analogous to those in the reference set, and to quantify
how different conditions in the set of interest are. The tools included
here help to identify conditions outside the rages of the reference set
with greater detail than in earlier implementations. These tools are
based on the methods proposed by Owens et
al. (2013) and presented in Cobos et
al. (2024).
To cite the methods implemented in this package use:
Cobos ME, Owens HL, Soberón J, Peterson AT (2024) Detailed multivariate comparisons of environments with mobility oriented parity. Frontiers of Biogeography 17: e132916. https://doi.org/10.21425/fob.17.132916
Big thanks to code contributor: Dave Slager.
To install the stable version of mop use:
install.packages("mop")Before installing the development version of mop, make
sure to obtain the compilation tools required: Rtools for
Windows, Xcode
for Mac, and ggc or similar compilers in Linux, see
examples here or here.
After that, you can install the development version of
mop from its GitHub repository
with:
# install.packages("remotes")
remotes::install_github("marlonecobos/mop")The following are basic examples of how to use the main function of the package. First, load the package and some example data.
# package
library(mop)
# data
## current conditions
reference_layers <- terra::rast(system.file("extdata", "reference_layers.tif",
package = "mop"))
## future conditions
layers_of_interest <- terra::rast(system.file("extdata",
"layers_of_interest.tif",
package = "mop"))
# plot the data
## variables to represent current conditions
terra::plot(reference_layers)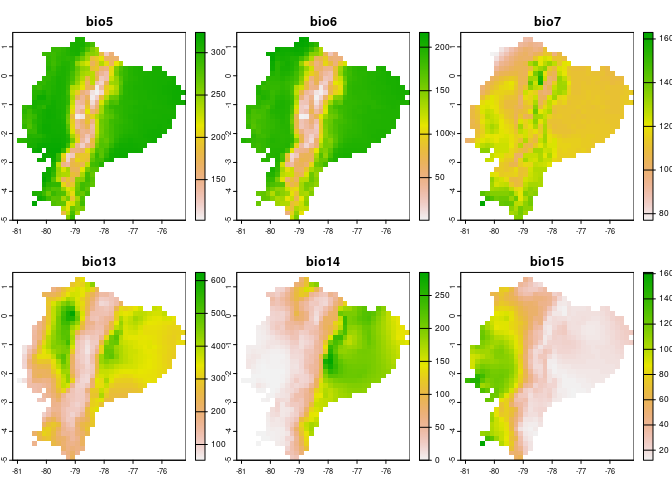
## variables to represent future conditions
terra::plot(layers_of_interest)
The code below helps to run analyses with all the details implemented
in the function. To see more basic options and what they imply, check
the function documentation with help(mop). Parallel
processing is allowed via arguments of this function.
# analysis
mop_basic_res <- mop(m = reference_layers, g = layers_of_interest,
type = "detailed", calculate_distance = TRUE,
where_distance = "all", distance = "euclidean",
scale = TRUE, center = TRUE)
#> | | | 0% | |=================================== | 50% | |======================================================================| 100%
# summary
summary(mop_basic_res)
#>
#> Summary of MOP resuls
#> ---------------------------------------------------------------------------
#>
#> MOP summary:
#> Values
#> type scale center calculate_distance distance percentage
#> 1 detailed TRUE TRUE TRUE euclidean 1
#> rescale_distance fix_NA N_m N_g
#> 1 FALSE TRUE 723 723
#>
#> Reference conditions
#> bio5 bio6 bio7 bio13 bio14 bio15
#> min -3.1625758 -2.8057083 -3.609429 -2.002072 -1.170877 -1.027498
#> max 0.6507328 0.8513394 3.710556 2.779598 2.351446 3.174031
#>
#>
#> Distances:
#> min mean max
#> 0.3003245 0.9373088 2.1788036
#>
#>
#> Non-analogous conditions (NAC):
#> Percentage = 0.566% of all conditions
#> Variables with NAC in 'simple' = 2
#>
#>
#> Detailed results were obtained, not shown here
# print results
mop_basic_res
#> MOP distances:
#> class : SpatRaster
#> dimensions : 39, 36, 1 (nrow, ncol, nlyr)
#> resolution : 0.1666667, 0.1666667 (x, y)
#> extent : -81.16667, -75.16667, -5, 1.5 (xmin, xmax, ymin, ymax)
#> coord. ref. : lon/lat WGS 84 (with axis order normalized for visualization)
#> source(s) : memory
#> varname : layers_of_interest
#> name : mop
#> min value : 0.3003245
#> max value : 2.1788036
#>
#> MOP basic:
#> class : SpatRaster
#> dimensions : 39, 36, 1 (nrow, ncol, nlyr)
#> resolution : 0.1666667, 0.1666667 (x, y)
#> extent : -81.16667, -75.16667, -5, 1.5 (xmin, xmax, ymin, ymax)
#> coord. ref. : lon/lat WGS 84 (with axis order normalized for visualization)
#> source(s) : memory
#> varname : layers_of_interest
#> name : mop
#> min value : 1
#> max value : 1
#>
#> MOP simple:
#> class : SpatRaster
#> dimensions : 39, 36, 1 (nrow, ncol, nlyr)
#> resolution : 0.1666667, 0.1666667 (x, y)
#> extent : -81.16667, -75.16667, -5, 1.5 (xmin, xmax, ymin, ymax)
#> coord. ref. : lon/lat WGS 84 (with axis order normalized for visualization)
#> source(s) : memory
#> varname : layers_of_interest
#> categories : n_variables
#> name : n_variables
#> min value : 1
#> max value : 2
#>
#> MOP detailed:
#> interpretation_combined:
#> values extrapolation_variables
#> 1 1e+01 bio5
#> 2 1e+02 bio6
#> 3 1e+03 bio7
#> 4 1e+04 bio13
#> 5 1e+05 bio14
#> 6 1e+06 bio15
#> ...
#>
#> towards_low_end:
#> class : SpatRaster
#> dimensions : 39, 36, 6 (nrow, ncol, nlyr)
#> resolution : 0.1666667, 0.1666667 (x, y)
#> extent : -81.16667, -75.16667, -5, 1.5 (xmin, xmax, ymin, ymax)
#> coord. ref. : lon/lat WGS 84 (with axis order normalized for visualization)
#> source(s) : memory
#> varnames : layers_of_interest
#> layers_of_interest
#> layers_of_interest
#> ...
#> names : bio5, bio6, bio7, bio13, bio14, bio15
#> min values : NaN, NaN, NaN, NaN, NaN, 1
#> max values : NaN, NaN, NaN, NaN, NaN, 1
#>
#> towards_high_end:
#> class : SpatRaster
#> dimensions : 39, 36, 6 (nrow, ncol, nlyr)
#> resolution : 0.1666667, 0.1666667 (x, y)
#> extent : -81.16667, -75.16667, -5, 1.5 (xmin, xmax, ymin, ymax)
#> coord. ref. : lon/lat WGS 84 (with axis order normalized for visualization)
#> source(s) : memory
#> varnames : layers_of_interest
#> layers_of_interest
#> layers_of_interest
#> ...
#> names : bio5, bio6, bio7, bio13, bio14, bio15
#> min values : 1, 1, 1, 1, 1, NaN
#> max values : 1, 1, 1, 1, 1, NaN
#>
#> towards_low_combined:
#> class : SpatRaster
#> dimensions : 39, 36, 1 (nrow, ncol, nlyr)
#> resolution : 0.1666667, 0.1666667 (x, y)
#> extent : -81.16667, -75.16667, -5, 1.5 (xmin, xmax, ymin, ymax)
#> coord. ref. : lon/lat WGS 84 (with axis order normalized for visualization)
#> source(s) : memory
#> varname : layers_of_interest
#> categories : extrapolation_variables
#> name : extrapolation_variables
#> min value : bio15
#> max value : bio15
#>
#> towards_high_combined:
#> class : SpatRaster
#> dimensions : 39, 36, 1 (nrow, ncol, nlyr)
#> resolution : 0.1666667, 0.1666667 (x, y)
#> extent : -81.16667, -75.16667, -5, 1.5 (xmin, xmax, ymin, ymax)
#> coord. ref. : lon/lat WGS 84 (with axis order normalized for visualization)
#> source(s) : memory
#> varname : layers_of_interest
#> categories : extrapolation_variables
#> name : extrapolation_variables
#> min value : bio5
#> max value : bio14Below are some example plots of the results that can be obtained from
analysis with mop.
# difference between set of conditions of interest and the reference set
terra::plot(mop_basic_res$mop_distances)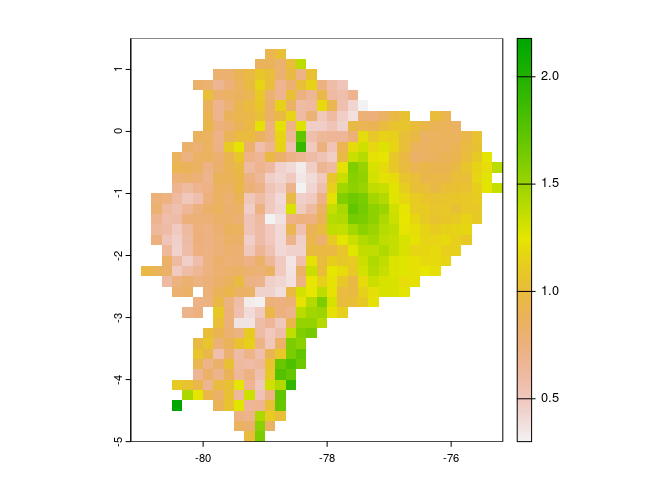
# basic identification of non-analogous results
terra::plot(mop_basic_res$mop_basic)
# how many variables have non-analogous conditions
terra::plot(mop_basic_res$mop_simple)
# combinations of variables with non-analogous conditions towards high values
terra::plot(mop_basic_res$mop_detailed$towards_high_combined)
# combinations of variables with non-analogous conditions towards low values
terra::plot(mop_basic_res$mop_detailed$towards_low_combined)