
mapmixture is an R package and Shiny app that enables
users to visualise admixture as pie charts on a projected map. It also
allows users to visualise admixture as traditional structure barplots or
facet barplots.
mapmixture requires R (>= 4.2) to be installed on
your system. Click here to download
the latest version of R for Windows.
Install the latest stable release from CRAN:
install.packages("mapmixture")Install the latest development version from GitHub:
# install.packages("devtools")
devtools::install_github("Tom-Jenkins/mapmixture")mapmixture() # main function
structure_plot() # plot traditional structure or facet barplot
scatter_plot() # plot PCA or DAPC results
launch_mapmixture() # launch mapmixture Shiny appJenkins TL (2024). mapmixture: an R package and web app for spatial visualisation of admixture and population structure. Molecular Ecology Resources, 24: e13943. DOI: 10.1111/1755-0998.13943.
# Load package
library(mapmixture)
# Read in admixture file format 1
file <- system.file("extdata", "admixture1.csv", package = "mapmixture")
admixture1 <- read.csv(file)
# Read in coordinates file
file <- system.file("extdata", "coordinates.csv", package = "mapmixture")
coordinates <- read.csv(file)
# Run mapmixture
map1 <- mapmixture(admixture1, coordinates, crs = 3035)
# map1
# Load packages
library(mapmixture)
library(rnaturalearthhires)
# Install rnaturalearthhires package using:
# install.packages("rnaturalearthhires", repos = "https://ropensci.r-universe.dev", type = "source")
# Read in admixture file format 1
file <- system.file("extdata", "admixture1.csv", package = "mapmixture")
admixture1 <- read.csv(file)
# Read in coordinates file
file <- system.file("extdata", "coordinates.csv", package = "mapmixture")
coordinates <- read.csv(file)
# Run mapmixture
map2 <- mapmixture(
admixture_df = admixture1,
coords_df = coordinates,
cluster_cols = c("#f1a340","#998ec3"),
cluster_names = c("Group A","Group B"),
crs = 3035,
basemap = rnaturalearthhires::countries10[, c("geometry")],
boundary = c(xmin=-15, xmax=16, ymin=40, ymax=62),
pie_size = 1,
pie_border = 0.3,
pie_border_col = "white",
pie_opacity = 1,
land_colour = "#d9d9d9",
sea_colour = "#deebf7",
expand = TRUE,
arrow = TRUE,
arrow_size = 1.5,
arrow_position = "bl",
scalebar = TRUE,
scalebar_size = 1.5,
scalebar_position = "tl",
plot_title = "Admixture Map",
plot_title_size = 12,
axis_title_size = 10,
axis_text_size = 8
)
# map2
# Load package
library(mapmixture)
# Read in admixture file format 3
file <- system.file("extdata", "admixture3.csv", package = "mapmixture")
admixture3 <- read.csv(file)
# Read in coordinates file
file <- system.file("extdata", "coordinates.csv", package = "mapmixture")
coordinates <- read.csv(file)
# Run mapmixture
map3 <- mapmixture(admixture3, coordinates, crs = 3035)
# map3
# Load packages
library(mapmixture)
library(ggplot2)
# Read in admixture file format 1
file <- system.file("extdata", "admixture1.csv", package = "mapmixture")
admixture1 <- read.csv(file)
# Read in coordinates file
file <- system.file("extdata", "coordinates.csv", package = "mapmixture")
coordinates <- read.csv(file)
# Run mapmixture
map4 <- mapmixture(
admixture_df = admixture1,
coords_df = coordinates,
cluster_cols = c("#f1a340","#998ec3"),
cluster_names = c("Ancestry 1","Ancestry 2"),
crs = 4326,
boundary = c(xmin=-15, xmax=16, ymin=40, ymax=62),
pie_size = 1,
)+
# Add additional label to the map
annotate("label",
x = -10,
y = 46.5,
label = "Atlantic Ocean",
size = 3,
)+
# Add additional text to the map
annotate("text",
x = 2.5,
y = 57,
label = "North Sea",
size = 3,
)+
# Adjust ggplot theme options
theme(
axis.title = element_text(size = 10),
axis.text = element_text(size = 8),
)+
# Adjust the size of the legend keys
guides(fill = guide_legend(override.aes = list(size = 5, alpha = 1)))
# map4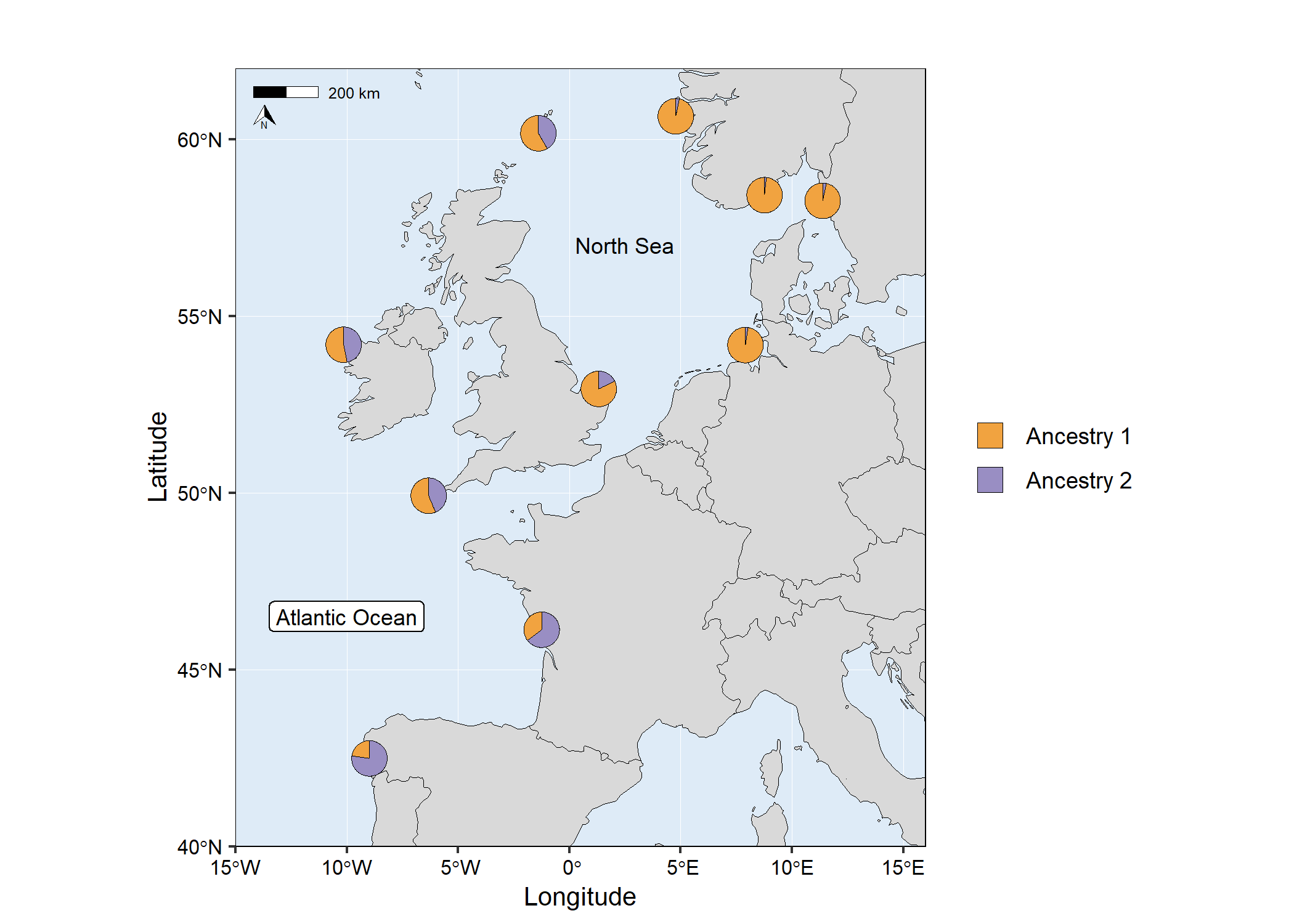
# Load packages
library(mapmixture)
library(ggplot2)
library(gridExtra)
# Read in admixture file format 1
file <- system.file("extdata", "admixture1.csv", package = "mapmixture")
admixture1 <- read.csv(file)
# Read in coordinates file
file <- system.file("extdata", "coordinates.csv", package = "mapmixture")
coordinates <- read.csv(file)
# Run mapmixture
map5 <- mapmixture(
admixture_df = admixture1,
coords_df = coordinates,
cluster_cols = c("#f1a340","#998ec3"),
cluster_names = c("Ancestry 1","Ancestry 2"),
crs = 4326,
boundary = c(xmin=-20, xmax=20, ymin=40, ymax=62),
pie_size = 1.3,
)+
# Adjust theme options
theme(
legend.position = "top",
plot.margin = margin(l = 10, r = 10),
)+
# Adjust the size of the legend keys
guides(fill = guide_legend(override.aes = list(size = 5, alpha = 1)))
# Traditional structure barplot
structure_barplot <- structure_plot(
admixture_df = admixture1,
type = "structure",
cluster_cols = c("#f1a340","#998ec3"),
site_dividers = TRUE,
divider_width = 0.4,
site_order = c(
"Vigo","Ile de Re","Isles of Scilly","Mullet Peninsula",
"Shetland","Cromer","Helgoland","Flodevigen","Lysekil","Bergen"
),
labels = "site",
flip_axis = FALSE,
site_ticks_size = -0.05,
site_labels_y = -0.35,
site_labels_size = 2.2
)+
# Adjust theme options
theme(
axis.title.y = element_text(size = 8, hjust = 1),
axis.text.y = element_text(size = 5),
)
# Arrange plots
# grid.arrange(map5, structure_barplot, nrow = 2, heights = c(4,1))
# Load packages
library(mapmixture)
library(ggplot2)
library(gridExtra)
# Read in admixture file format 1
file <- system.file("extdata", "admixture1.csv", package = "mapmixture")
admixture1 <- read.csv(file)
# Read in coordinates file
file <- system.file("extdata", "coordinates.csv", package = "mapmixture")
coordinates <- read.csv(file)
# Run mapmixture
map6 <- mapmixture(
admixture_df = admixture1,
coords_df = coordinates,
cluster_cols = c("#f1a340","#998ec3"),
cluster_names = c("Ancestry 1","Ancestry 2"),
crs = 4326,
boundary = c(xmin=-20, xmax=20, ymin=40, ymax=62),
pie_size = 1.3,
)+
# Adjust theme options
theme(
legend.position = "top",
plot.margin = margin(l = 10, r = 10),
)+
# Adjust the size of the legend keys
guides(fill = guide_legend(override.aes = list(size = 5, alpha = 1)))
# Facet structure barplot
facet_barplot <- structure_plot(admixture1,
type = "facet",
cluster_cols = c("#f1a340","#998ec3"),
facet_col = 2,
ylabel = "Admixture proportions",
)+
theme(
axis.title.y = element_text(size = 10),
axis.text.y = element_text(size = 5),
strip.text = element_text(size = 6, vjust = 1, margin = margin(t=1.5, r=0, b=1.5, l=0)),
)
# Arrange plots
# grid.arrange(map6, facet_barplot, ncol = 2, widths = c(3,2))
The raster (TIFF) used in the example below was downloaded from
Natural Earth here.
You need to install the terra package to use this
feature. Currently, the basemap argument accepts a
SpatRaster or a sf object.
# Load packages
library(mapmixture)
library(terra)
# Create SpatRaster object
earth <- terra::rast("../NE1_50M_SR_W/NE1_50M_SR_W.tif")
# Read in admixture file format 1
file <- system.file("extdata", "admixture1.csv", package = "mapmixture")
admixture1 <- read.csv(file)
# Read in coordinates file
file <- system.file("extdata", "coordinates.csv", package = "mapmixture")
coordinates <- read.csv(file)
# Run mapmixture
map7 <- mapmixture(admixture1, coordinates, crs = 3035, basemap = earth)
# map7
The vector data (shapefile) used in the example below was downloaded from the Natural England Open Data Geoportal here.
# Load packages
library(mapmixture)
library(rnaturalearthhires)
library(ggplot2)
library(dplyr)
library(sf)
# Read in admixture file format 1
file <- system.file("extdata", "admixture1.csv", package = "mapmixture")
admixture1 <- read.csv(file)
# Read in coordinates file
file <- system.file("extdata", "coordinates.csv", package = "mapmixture")
coordinates <- read.csv(file)
# Parameters
crs <- 3035
boundary <- c(xmin=-11, xmax=13, ymin=50, ymax=60) |> transform_bbox(bbox = _, crs)
# Read in world countries from Natural Earth and transform to CRS
world <- rnaturalearthhires::countries10[, c("geometry")]
world <- st_transform(world, crs = crs)
# Read in Marine Conservation Zones shapefile
# Extract polygons for Western Channel, Offshore Brighton and Swallow Sand
# Transform to CRS
mczs <- st_read("../Marine_Conservation_Zones_England/Marine_Conservation_Zones___Natural_England_and_JNCC.shp", quiet = TRUE) |>
dplyr::filter(.data = _, MCZ_NAME %in% c("Western Channel", "Offshore Brighton", "Swallow Sand")) |>
st_transform(x = _, crs = crs)
# Run mapmixture helper functions to prepare admixture and coordinates data
admixture_df <- standardise_data(admixture1, type = "admixture") |> transform_admix_data(data = _)
coords_df <- standardise_data(coordinates, type = "coordinates")
admix_coords <- merge_coords_data(coords_df, admixture_df) |> transform_df_coords(df = _, crs = crs)
# Plot map and add pie charts
map8 <- ggplot()+
geom_sf(data = world, colour = "black", fill = "#d9d9d9", size = 0.1)+
geom_sf(data = mczs, aes(fill = "MCZs"), linewidth = 0.3)+
scale_fill_manual(values = c("yellow"))+
coord_sf(
xlim = c(boundary[["xmin"]], boundary[["xmax"]]),
ylim = c(boundary[["ymin"]], boundary[["ymax"]])
)+
add_pie_charts(admix_coords,
admix_columns = 4:ncol(admix_coords),
lat_column = "lat",
lon_column = "lon",
pie_colours = c("green","blue"),
border = 0.3,
opacity = 1,
pie_size = 0.8
)+
theme(
legend.title = element_blank(),
)
# map8
# Load packages
library(mapmixture)
library(ggplot2)
library(adegenet)
library(RColorBrewer)
library(gridExtra)
# Load example genotypes
data("dapcIllus")
geno = dapcIllus$a
# Change population labels
popNames(geno) = c("Pop1","Pop2","Pop3","Pop4","Pop5","Pop6")
# Region names
region_names <- rep(c("Region1", "Region2"), each = 300)
# Define colour palette
cols = brewer.pal(nPop(geno), "RdYlBu")
# Perform PCA
pca1 = dudi.pca(geno, scannf = FALSE, nf = 3)
# Percent of genetic variance explained by each axis
percent = round(pca1$eig/sum(pca1$eig)*100, digits = 1)
# Scatter plot with centroids and segments
scatter1 <- scatter_plot(
dataframe = pca1$li,
group_ids = geno$pop,
type = "points",
axes = c(1,2),
percent = percent,
colours = cols,
point_size = 2,
point_type = 21,
centroid_size = 2,
stroke = 0.1,
plot_title = "PCA coloured by group_ids"
)+
theme(
legend.position = "none",
axis.title = element_text(size = 8),
axis.text = element_text(size = 6),
plot.title = element_text(size = 10),
)
# Same as scatter1 but no segments and axis 1 and 3 are shown
scatter2 <- scatter_plot(
dataframe = pca1$li,
group_ids = geno$pop,
type = "points",
axes = c(1,3),
percent = percent,
colours = cols,
point_size = 2,
point_type = 21,
centroids = TRUE,
centroid_size = 2,
segments = FALSE,
stroke = 0.1,
plot_title = "PCA no segments and axis 1 and 3 shown"
)+
theme(
legend.position = "none",
axis.title = element_text(size = 8),
axis.text = element_text(size = 6),
plot.title = element_text(size = 10),
)
# Same as scatter1 but coloured by region
scatter3 <- scatter_plot(
dataframe = pca1$li,
group_ids = geno$pop,
other_group = region_names,
type = "points",
axes = c(1,2),
percent = percent,
colours = cols,
point_size = 2,
point_type = 21,
centroid_size = 2,
stroke = 0.1,
plot_title = "PCA coloured by other_group"
)+
theme(
legend.position = "none",
axis.title = element_text(size = 8),
axis.text = element_text(size = 6),
plot.title = element_text(size = 10),
)
# Scatter plot with labels instead of points
scatter4 <- scatter_plot(
dataframe = pca1$li,
group_ids = geno$pop,
type = "labels",
labels = rownames(pca1$li),
colours = cols,
size = 2,
label.size = 0.10,
label.padding = unit(0.10, "lines"),
plot_title = "PCA using labels instead of points"
)+
theme(
legend.position = "none",
axis.title = element_text(size = 8),
axis.text = element_text(size = 6),
plot.title = element_text(size = 10),
)
# Arrange plots
# grid.arrange(scatter1, scatter2, scatter3, scatter4)
# Load package
library(mapmixture)
# Launch Shiny app
launch_mapmixture()
# Tested with the following package versions:
# shiny v1.8.0 (important)
# shinyFeedback v0.4.0
# shinyjs v2.1.0
# shinyWidgets 0.8.4
# bslib 0.7.0
# colourpicker 1.3.0
# htmltools v0.5.8.1
# waiter 0.2.5https://tomjenkins.shinyapps.io/mapmixture/
# Load package
library(mapmixture)
# Admixture Format 1
file <- system.file("extdata", "admixture1.csv", package = "mapmixture")
admixture1 <- read.csv(file)
head(admixture1)
#> Site Ind Cluster1 Cluster2
#> 1 Bergen Ber01 0.9999 1e-04
#> 2 Bergen Ber02 0.9999 1e-04
#> 3 Bergen Ber03 0.9999 1e-04
#> 4 Bergen Ber04 0.9999 1e-04
#> 5 Bergen Ber05 0.9999 1e-04
#> 6 Bergen Ber06 0.9999 1e-04
# Admixture Format 2
file <- system.file("extdata", "admixture2.csv", package = "mapmixture")
admixture2 <- read.csv(file)
admixture2
#> Site Ind Cluster1 Cluster2
#> 1 Bergen Bergen 0.9675212 0.03247879
#> 2 Cromer Cromer 0.8217114 0.17828857
#> 3 Flodevigen Flodevigen 0.9843806 0.01561944
#> 4 Helgoland Helgoland 0.9761543 0.02384571
#> 5 Ile de Re Ile de Re 0.3529000 0.64710000
#> 6 Isles of Scilly Isles of Scilly 0.5632444 0.43675556
#> 7 Lysekil Lysekil 0.9661722 0.03382778
#> 8 Mullet Peninsula Mullet Peninsula 0.5316833 0.46831667
#> 9 Shetland Shetland 0.5838028 0.41619722
#> 10 Vigo Vigo 0.2268444 0.77315556
# Admixture Format 3
file <- system.file("extdata", "admixture3.csv", package = "mapmixture")
admixture3 <- read.csv(file)
admixture3
#> Site Ind Cluster1 Cluster2
#> 1 Bergen Bergen 1 0
#> 2 Cromer Cromer 1 0
#> 3 Flodevigen Flodevigen 1 0
#> 4 Helgoland Helgoland 1 0
#> 5 Ile de Re Ile de Re 0 1
#> 6 Isles of Scilly Isles of Scilly 1 0
#> 7 Lysekil Lysekil 1 0
#> 8 Mullet Peninsula Mullet Peninsula 1 0
#> 9 Shetland Shetland 1 0
#> 10 Vigo Vigo 0 1
# Coordinates
file <- system.file("extdata", "coordinates.csv", package = "mapmixture")
coordinates <- read.csv(file)
coordinates
#> Site Lat Lon
#> 1 Bergen 60.65 4.77
#> 2 Cromer 52.94 1.31
#> 3 Flodevigen 58.42 8.76
#> 4 Helgoland 54.18 7.90
#> 5 Ile de Re 46.13 -1.25
#> 6 Isles of Scilly 49.92 -6.33
#> 7 Lysekil 58.26 11.37
#> 8 Mullet Peninsula 54.19 -10.15
#> 9 Shetland 60.17 -1.40
#> 10 Vigo 42.49 -8.99