[!CAUTION] This package is now obsolete and it will be archived in the future. Users should move to using either moocore or mooplot.


Maintainer: Manuel López-Ibáñez
Contributors: Manuel López-Ibáñez, Marco Chiarandini, Carlos M. Fonseca, Luís Paquete, Thomas Stützle, and Mickaël Binois.
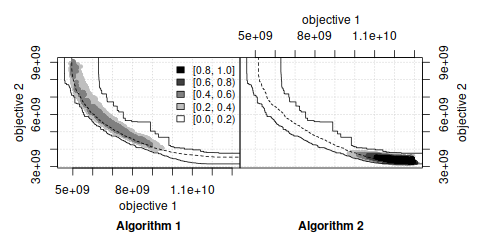
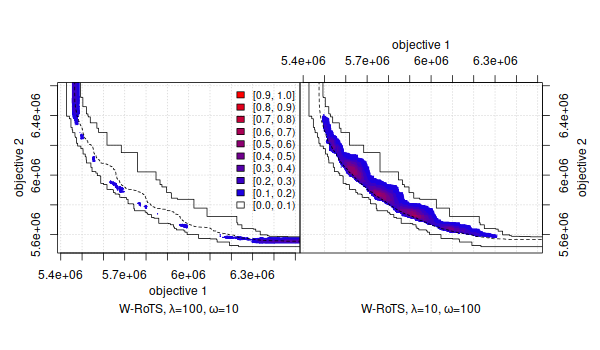
The empirical attainment function (EAF) describes the probabilistic distribution of the outcomes obtained by a stochastic algorithm in the objective space. This R package implements plots of summary attainment surfaces and differences between the first-order EAFs. These plots may be used for exploring the performance of stochastic local search algorithms for biobjective optimization problems and help in identifying certain algorithmic behaviors in a graphical way.
The corresponding book chapter [1] explains the use of these visualization tools and illustrates them with examples arising from practice.
In addition, the package provides functions for computing several quality metrics, such as hypervolume, IGD, IGD+, and epsilon.
Keywords: empirical attainment function, summary attainment surfaces, EAF differences, multi-objective optimization, bi-objective optimization, performance measures, performance assessment, graphical analysis, visualization.
Relevant literature:
The eaf package is implemented in R. Therefore, a basic knowledge of R is recommended to make use of all features.
The first step before installing the eaf package is to install R. Once R is installed in the system, there are two methods for installing the eaf package:
Install within R (automatic download, internet connection required). Invoke R, then
install.packages("eaf")Download the eaf package from CRAN (you may also need to download and install first the package modeltools), and invoke at the command-line:
R CMD INSTALL <package>where <package> is one of the three versions
available: .tar.gz (Unix/BSD/GNU/Linux), .tgz
(MacOS X), or .zip (Windows).
Search the R documentation if you need more help to install an R package on your system.
The code for computing the EAF is available as a C program, and it
does not require installing R or any R packages. Just download the package
source code, uncompress it, and look for the directory
src/eaf. The C code can be used to implement your own
visualizations instead of the visualizations provided by the
eaf package. Compiled executables for computing the EAF
can be found under system.file(package="eaf", "bin"). Other
useful executable programs can be found there.
The eaf package also contains two Perl scripts that may
allow you to generate standard plots without any R knowledge. See
inst/scripts/eafplot/ and
inst/scripts/eafdiff/ in the package source code. The
scripts use the eaf package internally to generate the
plots, and, hence, the eaf package must be installed
and working.
If you wish to be notified of bugfixes and new versions, please subscribe to the low-volume emo-list, where announcements will be made.
[ Download eaf package from CRAN ] [ Documentation ] [ Development version (GitHub) ]
If you wish to try the development version, you can install it by executing the following commands within the R console:
R> install.packages("devtools")
R> devtools::install_github("MLopez-Ibanez/eaf")Once the eaf package is installed, the following R commands will give more information:
library(eaf)
?eaf
?eafplot
?eafdiffplot
?read.data.sets
example(eafplot)
example(eafdiffplot) # This one takes some timeApart from the main R package, the source code contains the following
extras in the directory inst/ (after installation, these
files can be found at the directory printed by the R command
system.file(package="eaf")):
scripts/eafplot : Perl script to plot summary
attainment surfaces.scripts/eafdiff : Perl script to plot the differences
between the EAFs of two input sets.extdata/ : Examples of utilization of the above
programs. These are discussed in the corresponding book chapter [1].In addition, the source code contains the following under
src/: * src/eaf : This C program computes the
empirical attainment function in 2 or 3 dimensions. It is NOT required
by the other programs, but it is provided as a useful command-line
utility. This version is based on the original code written by Carlos M.
Fonseca available at http://www.tik.ee.ethz.ch/pisa/. A more recent
version is available at Prof. Fonseca’s
website. * src/mo-tools : Several tools for working
with multi-objective data.
For more information, consult the README files at each
subdirectory.
Thanks to rpy2, you
can use the eaf package from Python. A complete example
would be:
import os
## Uncomment this if you suffer from this bug in cffi 1.13.0
## https://bitbucket.org/rpy2/rpy2/issues/591/runtimeerror-found-a-situation-in-which-we
#os.environ['RPY2_CFFI_MODE'] = "API"
# Tested with rpy2 2.9.2-1 and 3.2.6
import numpy as np
from rpy2.robjects.packages import importr, isinstalled, PackageNotInstalledError
from rpy2.robjects import r as R
from rpy2.robjects import numpy2ri
from rpy2.robjects.vectors import StrVector
numpy2ri.activate()
from rpy2.interactive import process_revents
process_revents.start()
def install_rpackages(packages):
if not isinstance(packages, list):
packages = [ packages ]
utils = importr('utils') # import R's utility package
# Selectively install what needs to be installed.
names_to_install = [x for x in packages if not isinstalled(x)]
if len(names_to_install) > 0:
print(f"Installing packages: {names_to_install}")
utils.install_packages(StrVector(names_to_install), repos = "https://cloud.r-project.org", verbose=True)
try:
eaf = importr("eaf")
except PackageNotInstalledError as e:
install_rpackages("eaf")
eaf = importr("eaf") # Retry after install
path = R('system.file(package="eaf", "extdata")')[0] + "/"
alg1 = eaf.read_data_sets_(path + "ALG_1_dat.xz")
alg1 = np.asarray(alg1)
alg2 = np.asarray(eaf.read_data_sets_(path + "ALG_2_dat.xz"))
eaf.eafplot(alg1[:, 0:2], sets=alg1[:,2])
input("Press ENTER to see next plot: ")
eaf.eafdiffplot(alg1, alg2, title_left="A", title_right="B")This software is Copyright (C) 2011-2021 Carlos M. Fonseca, Luís Paquete, Thomas Stützle, Manuel López-Ibáñez and Marco Chiarandini.
This program is free software (software libre); you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation; either version 2 of the License, or (at your option) any later version.
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
IMPORTANT NOTE: Please be aware that the fact that this program is released as Free Software does not excuse you from scientific propriety, which obligates you to give appropriate credit! If you write a scientific paper describing research that made substantive use of this program, it is your obligation as a scientist to (a) mention the fashion in which this software was used in the Methods section; (b) mention the algorithm in the References section. The appropriate citation is:
Moreover, as a personal note, I would appreciate it if you would
email manuel.lopez-ibanez@manchester.ac.uk with citations
of papers referencing this work so I can mention them to my funding
agent and tenure committee.